Inclusion and ethics
Informed written consent was given by all participants or their legal representatives, for their medical data and biomaterials to be used for scientific research. The study was conducted in accordance with the Declaration of Helsinki44 and was approved by the local ethical committees of each participating center (ADC: AD CSF biobank METC no. 2017.315, SPIN: COLLECTION 16/2013, DDBB/BMDB Validation: H-23001553, UNIPG αS-SAA: Comitato Etico Aziende Sanitarie Regione Umbria 19369/AV and 20942/21/OV, BIODEM/UAntwerp Autopsy: EC of ZNA (approval no. 4363) and EC UZA / UAntwerp (14/12/130), YUHS DaT-PET: review board of Severance Hospital (IRB No. 4-2018-0944 and IRB No. 4-2024-0877)).
Immunoassay development and optimization
Immunoassay development was performed on the SimplePlex Ella platform (ProteinSimple) in CSF and the Quanterix Simoa platform (Quanterix) in CSF, plasma and serum. Both assays are based on a sandwich ELISA setup and were optimized for customizable assay parameters, that is, antibody combinations and concentrations, buffer conditions, sample dilution and volume. The Simoa assay was additionally optimized for addition of helper beads, incubation steps and times, and substrate concentration. A more detailed description of the assay optimization steps is given in Supplementary Methods 1.
Analytical validation
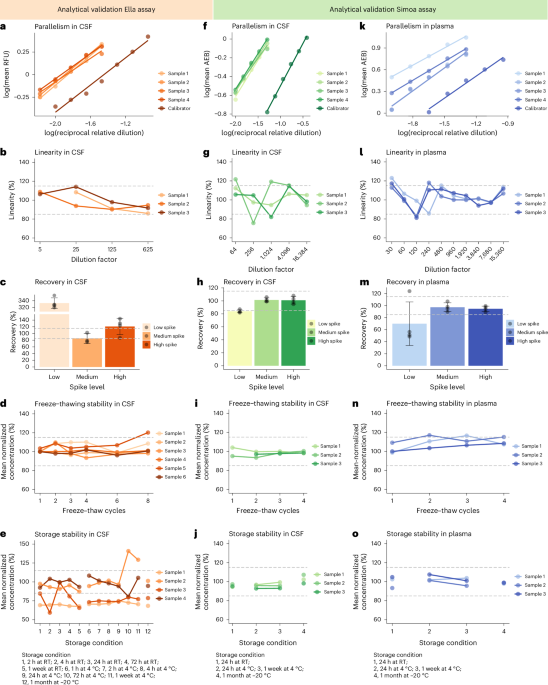
Analytical validations of the in-house-developed immunoassays were performed according to consensus guidelines25. Both assays were validated for lower limit of detection, precision, parallelism, dilution linearity, recovery and sample stability for the use in CSF (Supplementary Methods 2), and the Simoa assay was additionally validated for the use in plasma and serum (Supplementary Methods 3). Coefficients of variations of <20% were considered acceptable, and for parallelism, dilution linearity, recovery and sample stability, the accepted range of deviation of the samples from the reference condition was 85% to 115%.
Study population
We included participants from six different cohorts: ADC16 Discovery (CTRL, n = 50; DLB, n = 41; AD, n = 51), a clinical multicenter validation cohort from the ADC and SPIN45 (ADC/SPIN Validation; CTRL, n = 108; DLB; n = 106; MCI-A+, n = 101; AD dementia, n = 104), a DLB/PD cohort from the DDBB/BMDB (DDBB/BMDB Validation; CTRL, n = 44; DLB, n = 46; PD, n = 37; AD; n = 52), a biologically confirmed cohort from the University of Perugia, which consisted of participants with αS-SAA measurements previously performed with the 24-h Amprion protocol12 (UNIPG αS-SAA; CTRL (negative αS-SAA), n = 65; AD continuum, n = 110 (MCI-AD, n = 61; pre-AD, n = 11; AD dementia, n = 38); PD + DLB, n = 78 (all αS-SAA positive; cognitively normal PD, n = 33; PD-MCI, n = 34; PDD/DLB, n = 11)), an autopsy-confirmed cohort from the Reference Center for Biological Markers of Dementia (BIODEM) and the neurobiobank of the Institute Born-Bunge of the University of Antwerp (BIODEM/UAntwerp Autopsy; CTRL, n = 30; pathologically confirmed DLB, n = 18; pathologically confirmed AD, n = 30) and a cohort with detailed DaT-PET imaging data from the Yonsei University Health System of which n = 90 (88.2%) presented with significant parkinsonism (YUHS DaT-PET; total, n = 102, DaT-PET normal/abnormal, n = 24/78), which did not include healthy controls. Of note, the ADC Discovery Cohort, ADC/SPIN Validation Cohort and the BIODEM/UAntwerp Autopsy Cohort were also included in our previous proteomics study16.
AD core CSF biomarkers (Aβ42 or Aβ42/40, pTau181 and tTau) were used to support the clinical diagnosis of AD (abnormal Aβ42 or Aβ42/40 ratio and abnormal pTau181) and to confirm underlying amyloid pathology in the MCI group (as defined by abnormal Aβ42 or Aβ42/40 ratio) in the ADC Discovery, ADC/SPIN Validation and DDBB/BMDB Validation. In the UNIPG αS-SAA, AD was defined based on abnormal Aβ42/40 ratio and abnormal pTau181 (ref. 46) and in the YUHS DaT-PET AD was defined based on cerebral amyloid deposition measured by Florbetaben PET47. These markers were measured at each center with commercially available assays (ELISA INNOTEST Aβ1–42, hTAUAg, phospho-Tau(181 P) (Fujirebio-Europe): ADC, BIODEM-IBB/University of Antwerp, Danish Dementia Biobank; Aβ1–42, total-Tau, phospho-Tau181 Elecsys biomarker assays (Roche Diagnostics): ADC; Lumipulse G600 (Fujirebio-Europe): SPIN, University of Perugia). To determine biomarker abnormality, lab-specific cutoff values were applied (ADC48,49: Aβ42Innotest < 813 pg ml−1, pTauInnotest > 52 pg ml−1, tTauInnotest > 375 pg ml−1 or Aβ42Elecsys < 1,000 pg ml−1, pTauElecsys > 19 pg ml−1, tTauElecsys > 235 pg ml−1; SPIN50: Aβ42/40Lumipulse < 0.062, pTauLumipulse > 63 pg ml−1, tTauLumipulse > 456 pg ml−1; BIODEM/U Antwerp51: Aβ42Innotest < 638.5 pg ml−1, pTauInnotest > 56.5 pg ml−1, tTauInnotest > 296.5 pg ml−1; Danish Dementia Biobank52: Aβ42Innotest < 400 pg ml−1 (for measurements before 2013), <550 pg ml−1 (for measurements 2013–2018) or Aβ42Innotest < 1,016 pg ml−1 (for measurements between 2018 and July 2022), or Aβ42Elecsys < 1,030 pg ml−1 (for measurements after July 2022), pTauInnotest > 60 pg ml−1 (for patients younger than 60 years old) or >80 pg ml−1 (for patients older than 60 years old) before 2022 or pTauElecsys > 25 pg ml−1 from 2022 onward, tTauInnotest > 400 pg ml−1 before 2022 or tTauElecsys > 250 pg ml−1 from 2022 onward; University of Perugia53: Aβ42/40Lumipulse < 0.072, pTauLumipulse > 50 pg ml−1, tTauLumipulse > 392 pg ml−1). AD CSF biomarker measurements were available for subsets of the patients with DLB and PD: ADC Discovery, n = 38; ADC/SPIN Validation, n = 106; DDBB/BMDB Validation (PD), n = 14, DDBB/BMDB Validation (DLB), n = 32; UNIPG αS-SAA (CSF Ella), n = 55; UNIPG αS-SAA (CSF Simoa) Aβ42/40, n = 73; pTau and tTau, n = 75; UNIPG αS-SAA (Plasma Simoa) Aβ42/40, n = 50; pTau and tTau, n = 52; BIODEM/UAntwerp Autopsy, n = 13. In the YUHS DaT-PET, AD CSF biomarkers were available for all patients.
Information on DaT-Scan status was available for subsets of patients with DLB and PD; ADC Discovery (n DaT-Scan performed = 6), ADC/SPIN Validation (n DaT-Scan performed = 48) and DDBB/BMDB Validation (n DaT-Scan performed in DLB = 7; n DaT-Scan performed in PD = 19). Abnormal DaT-Scan could support the clinical diagnoses of 50/61 patients with DLB and 19/19 patients with PD. All patients in the YUHS DaT-PET underwent DaT-PET imaging (Supplementary Methods 4). Information on presence of DLB core features was available for subsets of the DLB group in ADC Discovery and ADC/SPIN Validation (Extended Data Table 6).
In all cohorts, participants underwent standard neurological and cognitive assessments and patients with DLB, PD, mild cognitive impairment and AD were diagnosed according to the respective diagnostic criteria6,7,46,54,55, or were defined according to international neuropathological examination guidelines for AD56 and DLB7,57 (BIODEM/UAntwerp Autopsy). Additionally, all patients with PD + DLB from the UNIPG αS-SAA, had positive CSF αS-SAA. Approximately 50% of the patients with AD in the UNIPG αS-SAA had positive CSF αS-SAA. Diagnoses in the YUHS DaT-PET were made based on clinical examination, neurological examination, AD CSF biomarkers and brain magnetic resonance imaging and included one or more etiologies for cognitive decline including AD, DLB, normal pressure hydrocephalus, vascular cognitive impairment, progressive supranuclear palsy and semantic variant primary progressive aphasia. Patients who exhibited parkinsonism or mild fluctuations and visual hallucinations that were not sufficient to be classified as DLB were categorized as possible LBD (pLBD). Controls in ADC Discovery and ADC/SPIN Validation were participants with subjective cognitive decline who did not present with any cognitive abnormalities during the diagnostic workup (ADC), or volunteers with normal neuropsychological scores (SPIN45). Controls in the DDBB/BMDB Validation were cognitively healthy individuals who were referred to diagnostic evaluation at the Danish Dementia Research Centre’s memory clinic but who were found without dementia or MCI. Controls of the UNIPG αS-SAA were cognitively normal individuals (n = 32) or individuals with MCI who had a normal AD CSF biomarker profile (n = 33) and negative CSF αS-SAA. Controls in the BIODEM/UAntwerp Autopsy were not pathologically confirmed and consisted of volunteers without any neurological or psychiatric diseases and did not present with any cognitive abnormalities58. Demographic characteristics of the included fluid cohorts are summarized in Extended Data Tables 1–3.
Immunohistochemistry was performed in postmortem brainstem tissue from pathology-confirmed controls (n = 4), patients with DLB (n = 4) and patients with PD(D) (n = 4). Brain tissue from patients with DLB and PD(D) was acquired from the Netherlands Brain Bank (http://brainbank.nl/) or from controls through the Normal Aging Brain Collection Amsterdam (http://nabca.eu/). The neuropathological diagnoses were established according to international guidelines of the Brain Net Europe II consortium (http://www.brainnet-europe.org/)56,59,60. Demographics and neuropathological characteristics of these donors are listed in Extended Data Table 8.
DDC measurements
DDC in CSF was measured with the in-house Ella and Simoa assays described above. In short, for measurements on the Ella system, 50 µl of CSF was measured in a twofold dilution. Inherent to the Ella technology, each sample was measured in triplicate. For measurements on the Simoa system, 100 µl of CSF were measured in a sixfold dilution. Each sample was measured in duplicate. For both assays, DDC concentrations were calculated with a four-parameter logistic curve, describing a calibration curve of recombinant human DDC. DDC concentrations in CSF were measured with our in-house-developed immunoassays in six different cohorts. DDC measurements on the Ella platform were performed in the ADC Discovery, ADC/SPIN Validation, BIODEM/UAntwerp Autopsy and UNIPG αS-SAA, and measurements on the Simoa platform were performed in the ADC Discovery, DDBB/BMDB Validation, UNIPG αS-SAA and YUHS DaT-PET. Ella measurements in the ADC/SPIN Validation were performed on Ella V4 cartridges, while the ADC Discovery, BIODEM/UAntwerp Autopsy and UNIPG αS-SAA were measured on the updated Ella V5 cartridges. We performed a Passing Bablok regression based on 50 samples from the ADC/SPIN Validation and identified that levels on the Ella V5 cartridges were about 10 pg ml−1 higher compared to measurements on the Ella V4 cartridges. The Passing Bablok regression resulted in the conversion formula as shown in equation (1):
$${\rm{Ella}}{\rm{V}}5=9.64+1.02\times {\rm{Ella}}{\rm{V}}4$$
(1)
This conversion formula has been applied to ADC/SPIN Validation before analysis to allow comparability between cohorts.
Additionally, PEA measurements of DDC in CSF were available for the ADC, SPIN and BIODEM/UAntwerp Autopsy from our previous study16.
Detection of DDC with immunohistochemistry
For DDC single stainings, 6-μm-thick formalin-fixed and paraffin embedded tissue sections from the brainstem, including the substantia nigra (at the level of the midbrain) and raphe nucleus (at the rostral level of the pons), were first deparaffinized using a xylene substitute and rehydrated in a series of ethanol with decreasing alcohol percentages. Next, antigen retrieval was performed for 30 min in citrate buffer (pH 6.0) in a steam cooker. Sections were washed in Tris-buffered saline (TBS) and incubated in TBS with 1% hydrogen peroxide (H2O2) for 30 min at room temperature to block endogenous peroxidase activity. Afterward, sections were washed with TBS at room temperature. To prevent nonspecific binding of the primary antibody, sections were blocked in TBS with 5% of elk milk powder and 0.1% Triton X-100 for 30 min. Subsequently, sections were incubated with primary anti-DDC antibody (1:400 dilution; AF3564, R&D Systems, detection antibody from DDC Ella and Simoa immunoassays), diluted in TBS overnight at 4 °C. The following day, sections were washed with TBS at room temperature and incubated for 30 min with ImmPRESS horse anti-goat antibody, coupled to horseradish peroxidase (HRP; MP-7405, Vector). After washing with TBS-HCl, Vector SG (SK-4700, Vector) was applied to the sections as chromogen for 25 min. Lastly, sections were dehydrated and mounted using Entellan.
Triple labeling experiments were performed for DDC, tyrosine hydroxylase and phosphorylated Ser129 α-syn on 6-μm-thick formalin-fixed and paraffin embedded sections from the substantia nigra. After deparaffinization and antigen retrieval as described above, sections were subsequently incubated in a blocking buffer containing 3% normal donkey serum and 0.1% Triton X in TBS for 30 min. Afterward, sections were incubated with primary anti-tyrosine hydroxylase antibody (MAB318, 1:200 dilution, Chemicon) overnight at 4 °C. The following day, sections were washed and incubated for 30 min at room temperature with the Envision system, containing secondary anti-mouse antibody, coupled to HRP (K400111-2, Dako). Subsequently, sections were washed in Tris-HCl and labeled with Tyramid Alexa Fluor 568 (B40956, 1:100 dilution, Thermo Fisher Scientific) in Tris-HCl + 0.005% H2O2 for 20 min. After the sections were washed in TBS and the antigen retrieval and blocking were repeated, sections were incubated with primary phosphorylated Ser129 α-syn (EP1536Y, 1:300 dilution, Abcam) and DDC (1:100 dilution, AF3564, R&D Systems) antibodies for 48 h at 4 °C. The following day, the sections were washed with TBS and incubated with Alexa Fluor 647-labeled donkey anti rabbit (A-31573, 1:400 dilution, Thermo Fisher Scientific) and DAPI (1:1,000 dilution, Thermo Fisher Scientific) for 2 h at room temperature. Sections were washed with TBS and incubated with ImmPRESS horse anti-goat antibody, coupled to HRP (MP-7405, Vector) for 30 min at room temperature, then washed with Tris-HCl and labeled with Tyramid Alexa Fluor 488 (B40953, 1:100 dilution, Thermo Fisher Scientific) in Tris-HCl with 0.005% H2O2 for 20 min, and subsequently washed in TBS. Sections were mounted in Mowiol mounting solution (Sigma-Aldrich) using glass coverslips. Negative control stainings lacking primary antibodies were performed to control for background/autofluorescence levels and unspecific staining. Single labeling for each antibody included in multiple-labeling experiments were carefully examined to control whether immunoreactivity patterns were caused by possible cross-reactivity between antibodies.
Brightfield and confocal laser scanning microscopy
The immunostained sections were first examined qualitatively by means of light microscopy (Leica DM5000 B photo microscope). High-resolution images were captured with the Leica DFC450 camera (HC PL APO ×20 1.30-NA oil or HC PL APO ×63 1.40-NA 0.60 oil objective) and the LAS 4.0 software. Neuromelanin-positive neurons in the substantia nigra of sections immunostained for phosphorylated α-syn/TH/DDC/DAPI were imaged using the Leica TCS SP8 STED ×3 confocal laser scanning microscope (CSLM: Leica Microsystems). All images were acquired using a HC PL APO CS2 ×100 1.4-NA oil objective lens, with the resolution set to a pixel size of 20 nm × 20 nm. Gated hybrid detectors were used in counting mode. Sections were sequentially scanned for each fluorophore, by irradiation with a pulsed white light laser at different wavelengths. Images were taken as z-stacks to allow deconvolution and three-dimensional image analysis (0.6 µm z-stack, 0.15 µm z-steps). Autofluorescence in the 488-nm and 594-nm channels was measured and extracted from the images, and compared to sections stained without primary antibodies (negative controls) to assess background signal. Deconvolution on the image stacks was performed using the Huygens Professional software package (Scientific Volume Imaging), using the manufacturer-recommended settings and the Batch Processor to apply the same settings to all images. Images were adjusted for brightness/contrast in the same way using an ImageJ (National Institute of Health).
Statistics and reproducibility
All statistical analyses were performed with R Studio (version 4.0.5)61. Distribution of data was analyzed by visual examination and Shapiro–Wilk test (pastecs R package62, version 1.4.2). To achieve a normal distribution, DDC, Aβ42, pTau and tTau concentrations were log10-transformed, and MMSE scores were subtracted from 31 and subsequently log10-transformed. Differences of DDC concentrations across diagnostic groups were assessed by ANCOVA (car R package63, version 3.1.2), corrected for age and sex (and center in ADC/SPIN Validation) with Tukey’s post hoc test. The potential of DDC to separate DLB from the other diagnostic groups was determined through ROC analysis (pROC R package64, version 1.18.5). First, we performed a ROC analysis for a basic model including only age and sex as predictors, or models including only DDC as predictor (DDC only) and next evaluated the added value of DDC to the basic models. Cutoff values were calculated from uncorrected and untransformed CSF DDC concentrations based on the Youden index. Meta-analyses of SMDs of DDC levels were performed between DLB and PD and other diagnostic groups (meta R package65, version 8.0.1). In cohorts that were measured with the Ella assay only, measurements were transformed to Simoa measurements beforehand. Association of DDC levels with cognitive impairment was analyzed by linear regression corrected for age and sex (and center in ADC/SPIN Validation). In the DLB group, associations between DDC levels and presence of DLB core features and AD CSF biomarker status were assessed through ANCOVA, corrected for age and sex (and center in ADC/SPIN Validation). Association of CSF DDC levels with motor impairment was assessed in the PD + DLB group in the UNIPG αS-SAA and in the YUHS DaT-PET by linear regression corrected for age, sex, LEDD and disease duration. Association between CSF DDC and DaT-PET in the caudate and putamen was analyzed by linear regression corrected for age, sex and education in the YUHS DaT-PET, and corrected for multiple testing by applying false discovery rate correction using the Benjamini–Hochberg procedure with a significance threshold of q < 0.05. Lastly, correlations between DDC measurements by PEA and by our in-house-developed immunoassays were analyzed with Spearman’s correlation. To evaluate the relation between the developed Ella and Simoa assays, as well as Ella V4 and Ella V5 cartridge format, we performed a Passing Bablok regression (mcr R package66, version 1.3.3). All statistical tests were two tailed, and a significance level of α = 0.05 was accepted.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.