Ethics committee approval
The outbreak investigation, including sample collection, was conducted as part of an emergency outbreak response led by the DRC Ministry of Public Health, Hygiene and Social Welfare and was therefore exempt from ethical approval. However, permission to use anonymous data for publication was granted by the Ethics Committee of the Kinshasa School of Public Health (ESP-UNIKIN, ethics approval no. ESP/CE/69/2025).
Statistical analysis
No statistical methods were applied.
Outbreak investigation setting and case definition
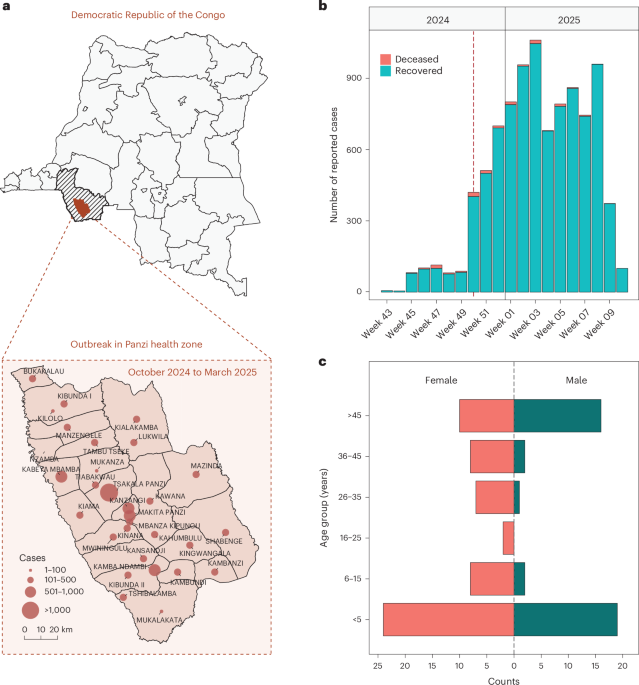
A multidisciplinary outbreak investigation team from the INRB traveled to Panzi on 8 December 2024 to collect specimens and epidemiological data from suspected individuals together with the local health team. Recruitment was unselected and conducted on a first-come-first-served basis on the basis of clinical attendance. Sex was determined on the basis of physical examination and registered in the medical record; gender was not collected. The outbreak investigation team included epidemiologists, entomologists, laboratory technicians, microbiologists and an anatomopathologist. Clinical samples were collected before treatment, between epidemiological weeks 49 and 51 of 2024 (early and mid-December), from individuals who met the following syndromic case definition: persons living in Panzi from September to December 2024 who presented to a health center with at least one of the signs or symptoms: fever, cough, fatigue and runny nose with or without chills, headache, dyspnea, malnutrition, pallor or body aches18.
Anthropometric measurements
Anthropometric parameters were collected by medical doctors from the INRB along with local nurses. Assessments were conducted following the DRC national protocol, which aligns with the WHO standards. Children aged <5 years were weighed using a salter scale (0–25 kg), while children aged >5 years and adults were weighed with standard scales. Height was measured using a wooden stadiometer (lying down for children aged 0–2 years and standing for those aged 3–5 years). Mid-upper arm circumference was measured in children aged <5 years.
Shipment of samples to the INRB, Kinshasa
Samples were shipped to the INRB in Kinshasa, DRC, in three separate batches on 7, 13 and 15 December 2024. The first batch consisted exclusively of blood samples, kept in a transport box with cold accumulators, collected from 12 suspected cases (laboratory IDs, 24INC-PAN01 to 12). The second batch included paired blood and oro/nasopharyngeal specimens, also in a transport box with cold accumulators, from 14 suspected cases (laboratory IDs, 24INC-PAN13 to 26). Both initial batches were collected by the local health team before the arrival of the outbreak investigation team from Kinshasa. The third and last batch—collected by the outbreak investigation team—comprised blood and oro/nasopharyngeal specimens from 82 suspected cases (laboratory IDs, 24INC-PAN27 to 108), among which 53 provided 79 blood cultures in BacT/ALERT (bioMérieux). Blood and oro/nasopharyngeal specimens collected by the outbreak investigation team were stored in liquid nitrogen, while BacT/ALERT bottles were kept at room temperature.
RDTs for P. falciparum detection
RDTs for P. falciparum were performed on capillary blood samples using the First Response Malaria Antigen P. falciparum card test (Premier Medical), designed to detect P. falciparum-specific histidine-rich protein 2 (HRP2) antigen. Tests were carried out according to the manufacturer’s instructions.
Blood cultures
Blood samples were collected by venipuncture using a blood collection adapter equipped with a sterile butterfly needle. For adults (as defined by the manufacturer as individuals aged ≥15 years), 2 × 10 ml blood was inoculated into aerobic BacT/ALERT bottles (bioMérieux), while for children (<15 years), a single sample of 1–4 ml blood was collected into a pediatric Bact/ALERT bottle (bioMérieux). In total, 79 aerobic BacT/ALERTs were collected from 53 cases, comprising 26 patients aged ≥15 years and 27 pediatric patients (<15 years). Inoculated BacT/ALERTs were shipped to the INRB, Kinshasa, at room temperature and then incubated at 35 °C for 5–7 days. BacT/ALERTs were checked daily for microbial growth by visual inspection of the colorimetric growth indicator.
Once growth was detected, Gram staining was performed, followed by subculture into MacConkey and 5% Sheep Blood agar (Difco). After 24 h incubation at 35 °C, phenotypic identification was performed using standard biochemical methods and confirmed with the VITEK 2 COMPACT System (bioMérieux). Antimicrobial susceptibility testing was performed on bacterial isolates following the Clinical and Laboratory Standards Institute guidelines using disk diffusion (Neo-Sensitabs, Rosco). For ciprofloxacin and azithromycin, the minimal inhibitory concentrations were determined using the E-test macromethod (bioMérieux, Oxoid). Extended-spectrum β-lactamase was assessed according to Clinical and Laboratory Standards Institute guidelines.
Processing of oro/nasopharyngeal swabs, and real-time multiplex PCR
Viral nucleic acid was extracted from 200 µl viral transport medium (3 ml) containing both oropharyngeal and nasopharyngeal swab specimens using the QIAmp Viral RNA mini kit (Qiagen), following the manufacturer’s instructions. Specimens were first tested using the Centers for Disease Control and Prevention (CDC) Influenza SARS-CoV-2 multiplex real-time PCR assay (catalog no. FluSC2PPB-RUO), followed by the FTD Respiratory Pathogens 21 assay (Fast Track Diagnostics), which allows the simultaneous detection of multiple respiratory pathogens such as influenza and parainfluenza viruses, RSV, human coronaviruses (excluding SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-2), human picornaviruses, metapneumoviruses, adenovirus and bocavirus, also following the manufacturer’s instructions. PCR testing was performed using the ABI 7500 Fast Real-Time PCR Systems (Life Technologies).
Blood collection and direct molecular testing
Blood was collected by venipuncture in EDTA tubes. Specimens were initially processed using the BioFire FilmArray System, an automated platform developed by BioFire Defense. This system extracts nucleic acids from inactivated whole blood samples and performs multiplex PCR with reverse transcription (RT–PCR) for simultaneous detection of pathogens in approximately 50 min. For this study, we used the BioFire Global Fever Panel (for Research Use Only (RUO)), which is able to detect 19 pathogens, including bacteria (Bacillus anthracis, Francisella tularensis, Leptospira spp., Salmonella enterica serovar Typhi and Paratyphi A and Yersinia pestis), viruses (Chikungunya virus, Crimean–Congo hemorrhagic fever virus, Dengue virus (serotypes 1–4), Ebolavirus spp., Marburgvirus, West Nile virus, Yellow fever virus and Zika virus) and protozoa (Plasmodium falciparum, Plasmodium spp., Plasmodium vivax/ovale and Leishmania spp.) (BioFire Defense). We also tested blood samples using the Biofire BioThreat panel (RUO), which is designed for detecting 16 biothreat pathogens and toxins such as filoviruses (Ebola virus and Marburg virus), mosquito-borne viruses (Eastern equine encephalitis virus, Venezuelan equine encephalitis virus and Western equine encephalitis virus), orthopoxviruses (mpox virus and Variola virus), bacteria (Bacillus anthracis, Brucella melitensis, Burkholderia mallei/pseudomallei, Coxiella burnetii, Francisella tularensis, Rickettsia prowazekii and Yersinia pestis) and toxin-encoding genes (Clostridium botulinum and Ricinus communis) (BioFire Defense).
Blood sample processing for real-time multiplex PCR and metagenomic sequencing
Total nucleic acid was extracted from 200 µl inactivated whole blood using the RADI universal DNA/RNA extraction kit (KH Medical), following the manufacturer’s protocol. Real-time PCR assays using TaqMan Microbial Array card (Blood/Cerebrospinal fluid Tier 1 v2; Thermo Fisher Scientific) were conducted on QuantStudio 7 Real-Time PCR system for detecting viruses (Dengue, Chikungunya, Zika, Yellow fever, West Nile, Rift Valley fever, Crimean–Congo hemorrhagic fever, Lassa, Nipah, Human parechovirus and Rubella), bacteria (Salmonella enterica Typhi and Paratyphi, Salmonella spp., Escherichia. coli, Shigella spp., Streptococcus spp., Listeria monocytogenes, Treponema pallidum and Rickettsia) and parasites (Plasmodium falciparum and P. vivax) associated with febrile syndromes. We also tested these extracts for filoviruses using the RealStar Filovirus Screen RT–PCR kit (Altona Diagnostics), following the manufacturer’s protocol.
For metagenomic sequencing, nucleic acid extracts were first subjected to random amplification using the sequence-independent, single-primer amplification (SISPA) method19. The purified SISPA products were subsequently used for both the Twist Comprehensive Viral Research (CVR) Panel probe-enrichment and metagenomic sequencing to enable identification of known and potentially novel pathogens. The Twist CVR Panel is a viral capture probe-based method designed to enrich viral sequences from 3,153 human and nonhuman viral species, including 15,488 distinct viral strains20.
Both SISPA and Twist probe-enriched products were used for library preparation. The libraries were prepared using the library sequencing amplicons V14 kit (SQK-LSK114; Oxford Nanopore Technologies), following the manufacturer’s protocol. Sequencing libraries were subsequently loaded onto R10 flow cells and run for 72 h on the GridION sequencer. In addition, total nucleic acids were processed using the Illumina RNA Prep with Enrichment Tagmentation (Illumina), which allows library preparation from degraded and low-yield RNA samples. The resulting libraries were then enriched using the Illumina Viral Surveillance Panel v2 kit (Illumina), following the manufacturer’s protocol. The final libraries were sequenced on an Illumina Iseq 100 system. Nuclease-free water control was included in each sequencing run during library preparation.
Bioinformatics analysis
Basecalling was performed using Dorado version 7.4.14. Demultiplexing and adapter trimming were done using the GridION built-in MinKNOW software version 24.06.15. FASTQ files were subsequently processed using a combination of different pipelines, including the atavide lite (https://github.com/linsalrob/atavide_lite) and wf-metagenomics (https://github.com/epi2me-labs/wf-metagenomics).
In brief, raw sequences were all processed using atavide-lite pipeline (https://doi.org/10.5281/zenodo.15356766), a modular metagenomics processing pipeline designed for large, complex and heterogeneous datasets. The pipeline integrates both read-based and assembly-based approaches, ensuring that taxonomic and functional inferences can be made from raw sequence data while also enabling genome reconstruction and binning. It supports both short-read paired-end datasets and long-read sequencing data, making it suitable for a broad range of sequencing technologies and study designs.
Raw fastq files were processed for quality control and adapter trimming using fastp21, which removes sequencing adapters and low-quality reads and prepares data for downstream processing. We mapped the high-quality reads to the human reference genome Genome Reference Consortium Human Build 38 (GRCh38) using minimap2 version 2.3022, and filtered out human reads using SAMtools version 1.22.123. Then, nonhuman reads were mapped against the UniRef50 reference database24 using MMseqs2 easy-taxonomy25. MMseqs2 easy-taxonomy parameters were optimized for a balance of sensitivity and speed, and included a 7-mer double-match prefilter with compositional bias correction, low-complexity masking with TANTAN and default similarity thresholds (k-score ~95) that generate sufficient similar k-mers for accurate detection while maintaining computational efficiency. Candidate hits were further filtered by fast ungapped alignment and refined with vectorized Smith–Waterman alignments, after which taxonomic labels were assigned using a lowest common ancestor approach as described by Steinegger and Soding25. The distribution of MMseqs2 alignment E-values spanned a wide range, from substantial matches (minimum E-value of 0) to weak hits (maximum 0.9). The bulk of matches were strong, as reflected by a median of 6.2 × 10−18, with the average skewed upward (1.4 × 10−3) by a minority of higher E-values. These summary statistics indicate that most alignments were highly important, while a small fraction of weak matches contributed disproportionately to the mean. The taxonomies were reformatted with taxonkit (version 0.20.0)26 and merged into a single table.
Functional annotations were enriched by mapping the UniProt IDs from the MMseq2 output directly to proteins associated with BV-BRC subsystems (this was a 1:1 mapping and did not require thresholds or cutoffs). The number of reads that MMseqs2 reported mapped to each protein was counted, and the total was divided by the number of mapped reads to normalize the read counts.
For the assembly-based approaches, metagenomic data were assembled using MEGAHIT version 1.2.927, which by default uses a succinct de Bruijn graph approach with multiple k-mer sizes (21–141 in steps of 20), automatic memory optimization and conservative heuristics for contig pruning to balance assembly sensitivity, contiguity and computational efficiency. Metagenome-assembled genomes were reconstructed using VAMB version 5.0.428, a variational autoencoder-based approach that jointly encodes k-mer composition and differential coverage profiles of contigs into a low-dimensional latent space. By learning these compressed representations, VAMB effectively separates contigs originating from different microbial genomes and clusters them using a medoid-based algorithm. This deep learning approach has been shown to improve bin purity and completeness compared with conventional binning methods such as MetaBAT 229 or CONCOCT30, particularly in complex metagenomic datasets31. All analyses were run on the Setonix Supercomputer at Pawsey Supercomputer Centre and visualized using Google Colab Notebooks.
Ethics and inclusion statement
This research was conducted in close collaboration with local partners throughout all stages of the study, including conceptualization and study design, field implementation, intellectual property, data ownership and authorship of resulting publications. The inclusion of the national, provincial and local researchers was central to the project, ensuring that their expertise and contextual knowledge guided the multidisciplinary investigation team and supported the implementation of medical countermeasures.
The study addressed the public health problem that was defined by the Ministry of Public Health, Hygiene and Social Welfare of the DRC. Roles and responsibilities of collaborators were clearly defined before the start of the outbreak investigation to ensure effective coordination and implementation of activities. In addition, targeted capacity-building activities were implemented, including outbreak investigation training for researchers from Panzi and bioinformatics training for researchers from the INRB.
The research did not require any exceptional authorization, as all procedures complied with existing national regulations. Ethical approval for the study was obtained at the national level, ensuring adherence to ethical requirements. All laboratory and field activities were performed in compliance with recommended standards of biosafety and environmental protection. The research posed no risk of stigmatization, discrimination or incrimination of participants, and appropriate measures were in place to safeguard their confidentiality and well-being. Likewise, no specific security and safety risks, stigmatization, incrimination or discrimination were identified during the study.
No transfer of biological materials or related knowledge out of the country involved benefit-sharing considerations. Finally, the findings from our study were shared with local and regional health authorities, and all relevant contributors were appropriately acknowledged to recognize the existing body of work and strengthen the contextual understanding of our results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.